Data & code
Scientists should ensure others, including members of society, have free and open access to code, software, and data.1 In the case of my published research, I have endeavored to provide all data and code necessary to replicate the data analysis and figures/tables on which I based my scientific conclusions. I am guided and inspired by the open access, documentation, and provenance standards of the National Science Foundation’s EarthCube project and the Geoscience Papers of the Future initiative.
I’ve also attempted to make available on the Internet many other scripts and code to accomplish other scientific and non-scientific computing tasks. These are mostly in R and, with apologies to those without the $2,150 for a copy, MATLAB. Most of my code (not all of it pretty or well-annotated) can be found in repositories on GitHub. Because I am an oceanographer and biogeochemist, the scripts pertain primarily to the geosciences. Among these is a set of scripts for processing and analyzing bacterial production data obtained using the 3H-leucine incorporation method. (Some of the original BP code is Krista Longnecker’s.)
Data and code for published academic research projects
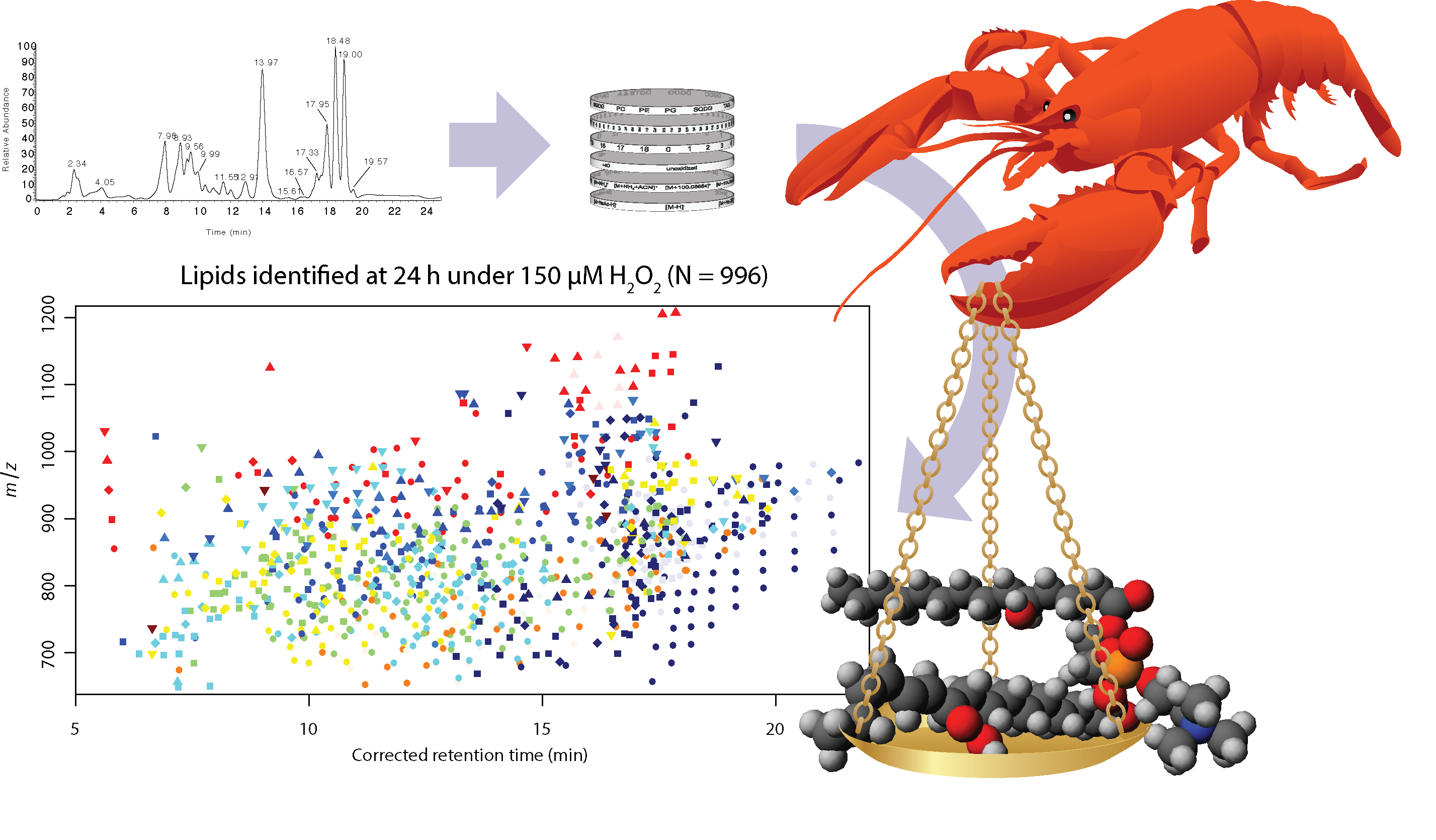 The LOBSTAHS software (Lipid and Oxylipin Biomarker Screening Through Adduct Hierarchy Sequences) is available at https://github.com/vanmooylipidomics/LOBSTAHS and is also part of Bioconductor. A manuscript describing LOBSTAHS has been published in Analytical Chemistry. Links, citable DOI, etc., are on the Publications page.
The LOBSTAHS software (Lipid and Oxylipin Biomarker Screening Through Adduct Hierarchy Sequences) is available at https://github.com/vanmooylipidomics/LOBSTAHS and is also part of Bioconductor. A manuscript describing LOBSTAHS has been published in Analytical Chemistry. Links, citable DOI, etc., are on the Publications page.
Data and code for other published projects can be downloaded using links on the Publications page. Once there, expand each entry using the icon; links to data and code appear at the end of each abstract.
Other software and code
Visit me on GitHub.
Some links for those interested in data, data science, and data visualization
» Federation of Earth Science Information Partners (ESIP). NASA/NOAA-funded initiative of the Foundation for Earth Science. A community which offers best practices and an amazing clearinghouse for new ideas on data, data stewardship, data discovery, and information technology in the earth sciences.
» The Biological and Chemical Oceanography Data Management Office (BCO-DMO). Also, some pointers for cocercing dates, times, geographical positions, and other data into an acceptable format for upload to BCO-DMO.
» Geoscience Papers of the Future Initiative. If you can, attend a training session.
» The Software Carpentry project.
» Some slides from a good talk at WHOI by C. Titus Brown on scientific workflows in Python. Talk was sponsored by WHOI’s Ocean Informatics initiative.
» Some modest advice about open-source software development: Slides I prepared for a January 27, 2017, workshop organized by WHOI’s Ocean Informatics initiative. Includes some advice on development of R packages for submission to Bioconductor, based on lessons learned from development of the LOBSTAHS package. Final few slides contain links to some Internet resources that cover package documentation and best practices for open-source software development. I’ve also posted slides from a newer version of this talk, which contain results of a brief survey on open-source development practices among postdocs & grad students at UW’s eScience Institute.